Explore Relationships and Ancestors
Meera Y. Patel, M.D.
2021-05-26
Source:vignettes/explore-relationships-and-ancestors.Rmd
explore-relationships-and-ancestors.RmdThere are 4 different types of relationships represented in the Concept Relationship table depending on whether they are hierarchical and if they define ancestry:
- Is Hierarchical & Does Not Define Ancestry
- Is Not Hierarchical & Defines Ancestry
- Is Hierarchical & Defines Ancestry
- Is Not Hierarchical & Does Not Define Ancestry
These relationship types can be queried directly in the Relationship table.
relationship_types <-
queryAthena(
conn = conn,
sql_statement =
"
SELECT DISTINCT
is_hierarchical,
defines_ancestry
FROM omop_vocabulary.relationship
")The relationship types are presented as boolean with the following definitions from the OMOP CDM v5.3.1 wiki:
is_hierarchical: Defines whether a relationship defines concepts into classes or hierarchies. Values are 1 for hierarchical relationship or 0 if not.
defines_ancestry: Defines whether a hierarchical relationship contributes to the concept_ancestor table. These are subsets of the hierarchical relationships. Valid values are 1 or 0.
relationship_types## # A tibble: 4 x 2
## is_hierarchical defines_ancestry
## <chr> <chr>
## 1 1 0
## 2 0 1
## 3 1 1
## 4 0 0Though a relationship that defines ancestry is by definition required to also be deemed hierarchical, non-hierarchical relationships that define ancestry appear to exist in the Relationship table.
SELECT DISTINCT
c.vocabulary_id AS vocabulary_id_1,
c.concept_class_id AS concept_class_id_1,
c2.vocabulary_id AS vocabulary_id_2,
c2.concept_class_id AS concept_class_id_2,
cr.relationship_id,
cr.invalid_reason
FROM omop_vocabulary.relationship r
INNER JOIN omop_vocabulary.concept_relationship cr
ON cr.relationship_id = r.relationship_id
INNER JOIN omop_vocabulary.concept c
ON cr.concept_id_1 = c.concept_id
INNER JOIN omop_vocabulary.concept c2
ON cr.concept_id_2 = c2.concept_id
WHERE
r.is_hierarchical = 0
AND r.defines_ancestry = 1
AND c.invalid_reason IS NULL
AND c2.invalid_reason IS NULL
outliers <-
queryAthena(sql_statement =
"
SELECT DISTINCT
c.vocabulary_id AS vocabulary_id_1,
c.concept_class_id AS concept_class_id_1,
c2.vocabulary_id AS vocabulary_id_2,
c2.concept_class_id AS concept_class_id_2,
cr.relationship_id,
cr.invalid_reason
FROM omop_vocabulary.relationship r
INNER JOIN omop_vocabulary.concept_relationship cr
ON cr.relationship_id = r.relationship_id
INNER JOIN omop_vocabulary.concept c
ON cr.concept_id_1 = c.concept_id
INNER JOIN omop_vocabulary.concept c2
ON cr.concept_id_2 = c2.concept_id
WHERE
r.is_hierarchical = '0'
AND r.defines_ancestry = '1'
AND c.invalid_reason IS NULL
AND c2.invalid_reason IS NULL
") %>%
mutate(`Vocabulary to Vocabulary` = paste0(vocabulary_id_1, " to ", vocabulary_id_2))The relationships indicate that these types of relationships only exist in the Drug Domain.
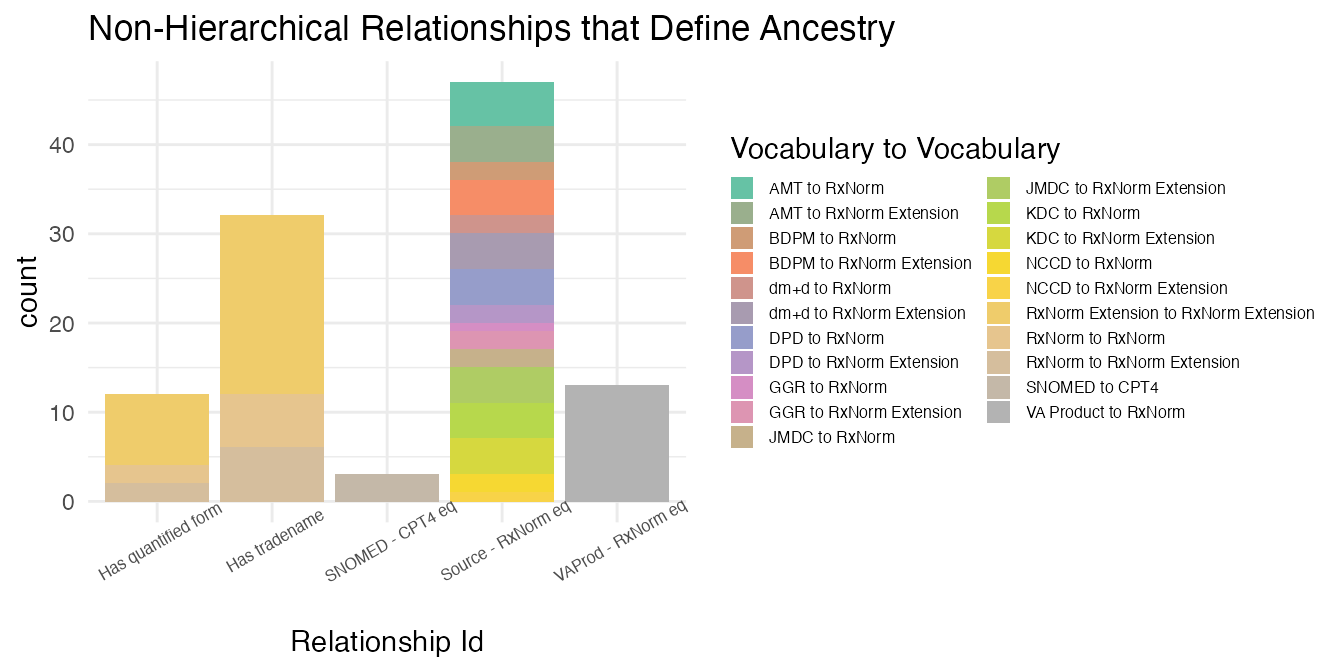
Full Review
For each of the 4 relationship types, any association with select controlled vocabularies SNOMED, LOINC, RxNorm, RxNorm Extension, HemOnc and ATC is derived. Relationships analyzed are filtered for those within each vocabulary system.
Methods
Relationships for each relationship type and target vocabulary is queried.
SELECT
c.vocabulary_id,
c.concept_class_id AS concept_class_id_1,
c2.concept_class_id AS concept_class_id_2,
r.relationship_id,
r.is_hierarchical,
r.defines_ancestry,
ca.min_levels_of_separation,
ca.max_levels_of_separation
FROM patelm9.@staging_table t
INNER JOIN omop_vocabulary.relationship r
ON t.is_hierarchical = r.is_hierarchical
AND t.defines_ancestry = r.defines_ancestry
INNER JOIN omop_vocabulary.concept_relationship cr
ON cr.relationship_id = r.relationship_id
INNER JOIN omop_vocabulary.concept c
ON cr.concept_id_1 = c.concept_id
INNER JOIN omop_vocabulary.concept c2
ON cr.concept_id_2 = c2.concept_id
INNER JOIN omop_vocabulary.concept_ancestor ca
ON ca.ancestor_concept_id = cr.concept_id_1
AND ca.descendant_concept_id = cr.concept_id_2
WHERE
c.invalid_reason IS NULL
AND c2.invalid_reason IS NULL
AND c.vocabulary_id IN ('@target_vocabulary')
AND c2.vocabulary_id IN ('@target_vocabulary')
AND cr.invalid_reason IS NULL
# Target vocabularies
target_vocabularies <- c("SNOMED", "LOINC",
"RxNorm", "RxNorm Extension",
"HemOnc", "ATC")
# Looping over each relationship type
output <- list()
for (i in 1:nrow(relationship_types)) {
relationship_type <-
relationship_types %>%
filter(row_number() == i)
conn <- connectAthena()
staging_table <- pg13::write_staging_table(conn = conn,
schema = "patelm9",
data = relationship_type,
drop_existing = TRUE,
drop_on_exit = FALSE)
output[[i]] <- list()
# Loop over each target vocabulary by relationship type
for (j in seq_along(target_vocabularies)) {
target_vocabulary <- target_vocabularies[j]
sql_statement <-
SqlRender::render(
[1234 chars quoted with '"'],
staging_table = staging_table,
target_vocabulary = target_vocabulary
)
output[[i]][[j]] <-
queryAthena(sql_statement = sql_statement)
names(output[[i]])[j] <- target_vocabulary
}
pg13::drop_all_staging_tables(conn = conn,
schema = "patelm9")
dcAthena(conn = conn)
}
# Add names to output
relationship_type_labels <-
c("Is Hierarchical & Does Not Define Ancestry",
"Is Not Hierarchical & Defines Ancestry",
"Is Hierarchical & Defines Ancestry",
"Is Not Hierarchical & Does Not Define Ancestry")
names(output) <- relationship_type_labels
R.cache::saveCache(object = output,
dirs = "chariot/vignettes/explore-relationships-and-ancestors/",
key = list("output")
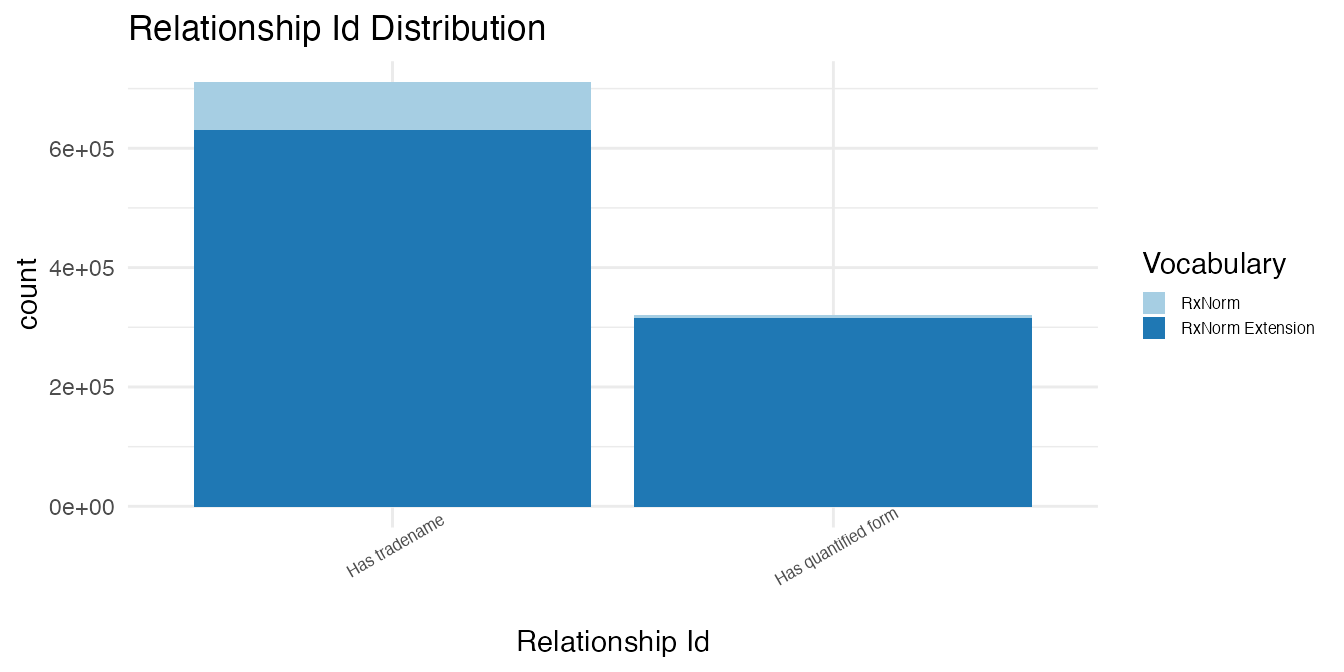
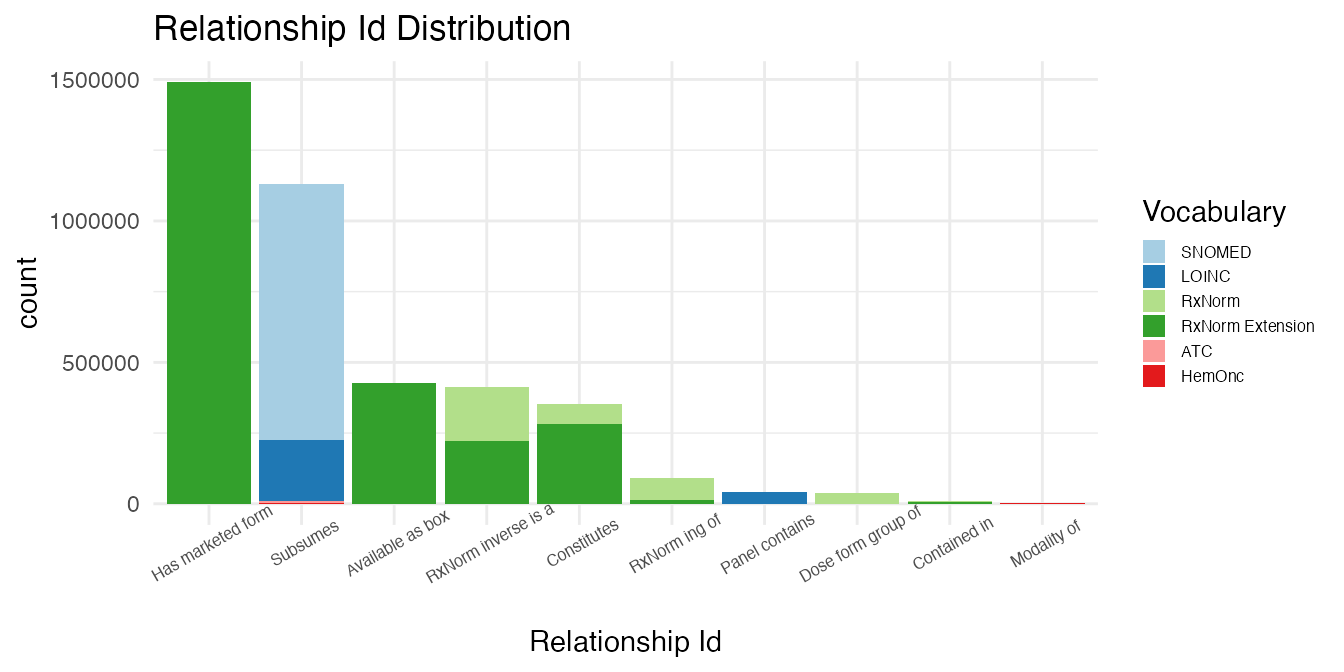
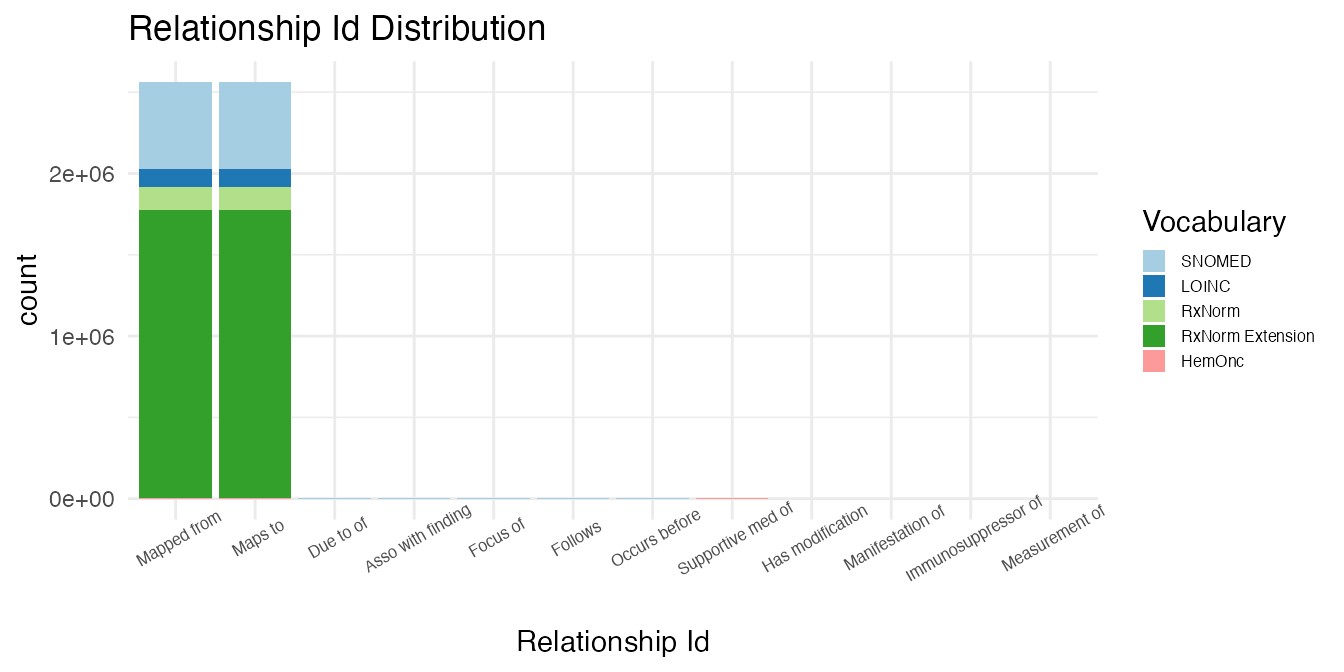
)The relationships are counted by relationship type.
# Bind outputs by relationship type
output2_a <-
output %>%
map(bind_rows) %>%
# Rename target fields for aesthetics in plots
map(function(x) x %>% rename(Vocabulary = vocabulary_id)) %>%
# Add relationship_id count
map(group_by,relationship_id) %>%
map(mutate, count = length(relationship_id)) %>%
map(ungroup) %>%
# Convert controlled vocabularies to factors
map(mutate_at, vars(Vocabulary), ~ factor(.,
levels = c("SNOMED",
"LOINC",
"RxNorm",
"RxNorm Extension",
"ATC",
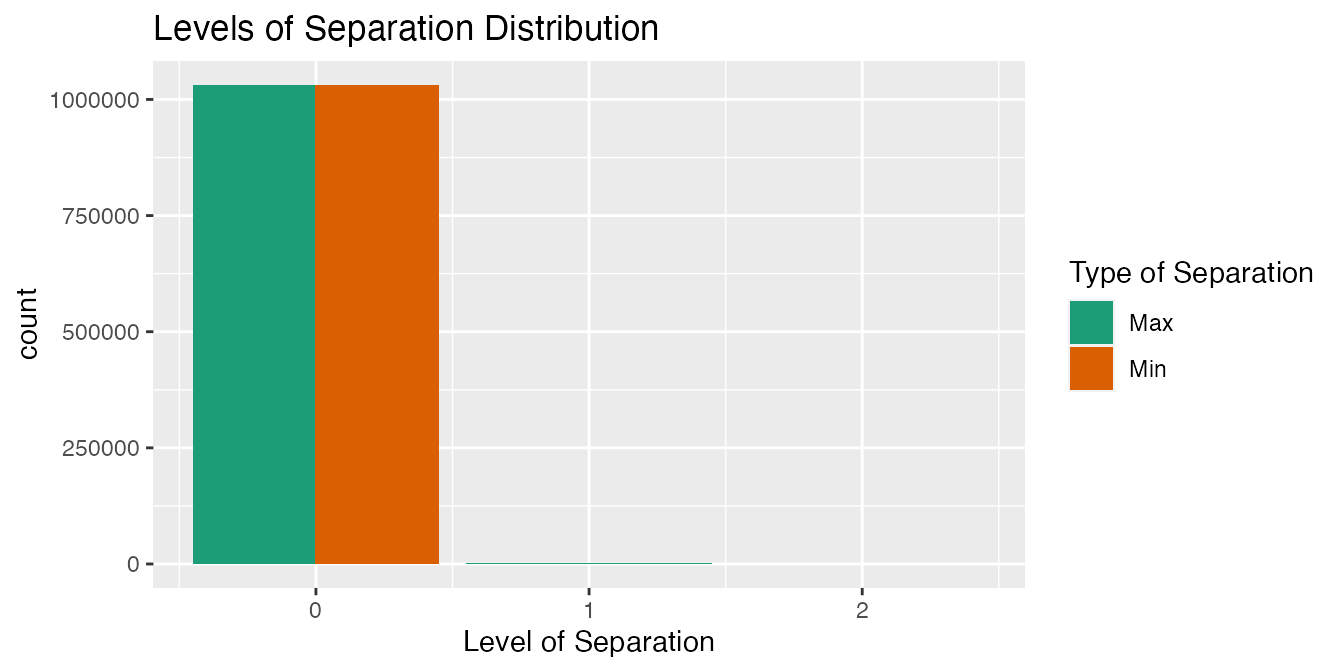
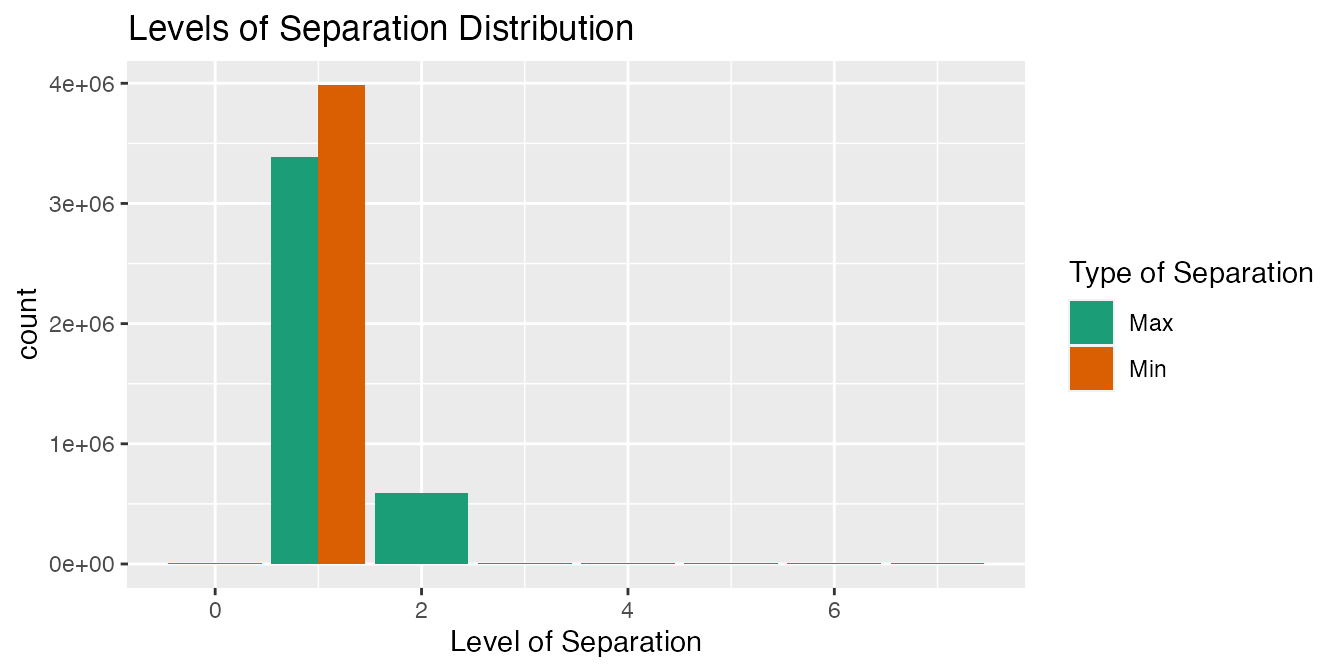
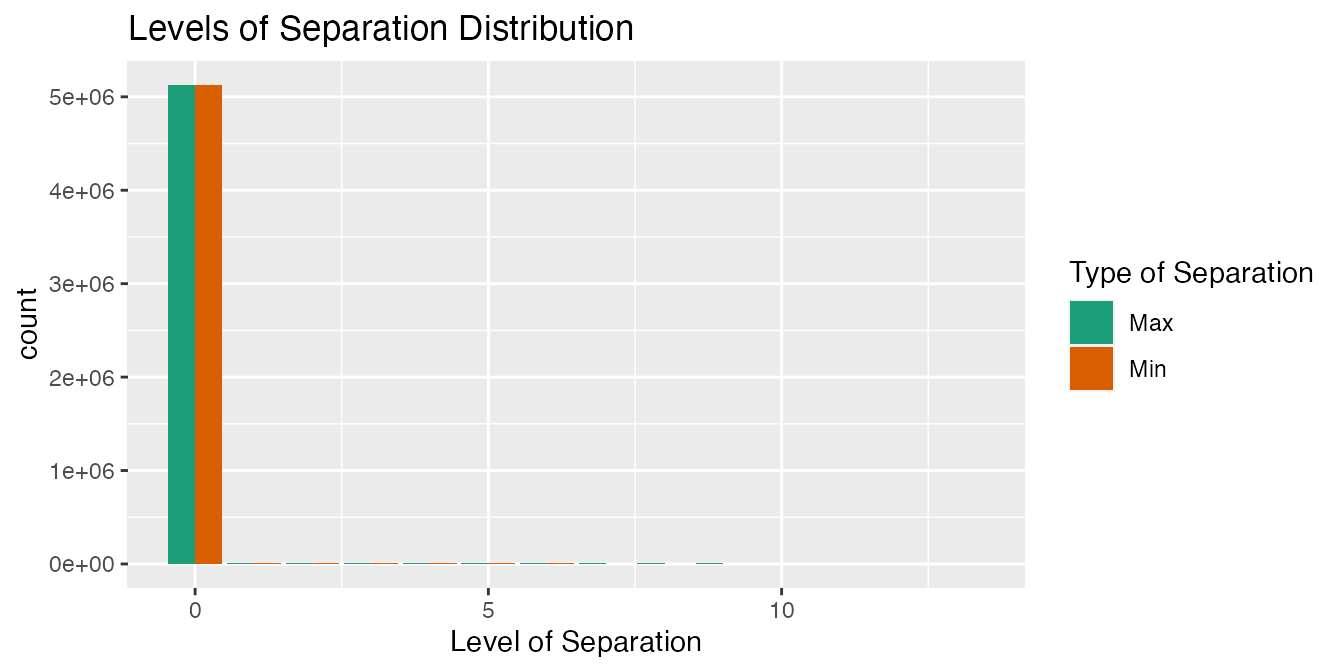
"HemOnc")))The distribution of max_levels_of_separation and min_levels_of_separation is also assessed by relationship type.
output2_b <-
output %>%
map(bind_rows) %>%
map(function(x) x %>%
pivot_longer(cols = c(min_levels_of_separation,
max_levels_of_separation),
names_to = "Type of Separation",
values_to = "level_of_separation")) %>%
map(function(x) x %>%
mutate_at(vars(`Type of Separation`),
function(y) case_when(y %in% c("max_levels_of_separation") ~ "Max",
TRUE ~ "Min")))Results
Hierarchical & Does Not Define Ancestry
There are zero relationships that are hierarchical, but do not define ancestry in the select controlled vocabularies.